Python insert numpy array into sqlite3 database
Question:
I’m trying to store a numpy array of about 1000 floats in a sqlite3 database but I keep getting the error “InterfaceError: Error binding parameter 1 – probably unsupported type”.
I was under the impression a BLOB data type could be anything but it definitely doesn’t work with a numpy array. Here’s what I tried:
import sqlite3 as sql
import numpy as np
con = sql.connect('test.bd',isolation_level=None)
cur = con.cursor()
cur.execute("CREATE TABLE foobar (id INTEGER PRIMARY KEY, array BLOB)")
cur.execute("INSERT INTO foobar VALUES (?,?)", (None,np.arange(0,500,0.5)))
con.commit()
Is there another module I can use to get the numpy array into the table? Or can I convert the numpy array into another form in Python (like a list or string I can split) that sqlite will accept? Performance isn’t a priority. I just want it to work!
Thanks!
Answers:
This works for me:
import sqlite3 as sql
import numpy as np
import json
con = sql.connect('test.db',isolation_level=None)
cur = con.cursor()
cur.execute("DROP TABLE FOOBAR")
cur.execute("CREATE TABLE foobar (id INTEGER PRIMARY KEY, array BLOB)")
cur.execute("INSERT INTO foobar VALUES (?,?)", (None, json.dumps(np.arange(0,500,0.5).tolist())))
con.commit()
cur.execute("SELECT * FROM FOOBAR")
data = cur.fetchall()
print data
data = cur.fetchall()
my_list = json.loads(data[0][1])
You could register a new array data type with sqlite3:
import sqlite3
import numpy as np
import io
def adapt_array(arr):
"""
http://stackoverflow.com/a/31312102/190597 (SoulNibbler)
"""
out = io.BytesIO()
np.save(out, arr)
out.seek(0)
return sqlite3.Binary(out.read())
def convert_array(text):
out = io.BytesIO(text)
out.seek(0)
return np.load(out)
# Converts np.array to TEXT when inserting
sqlite3.register_adapter(np.ndarray, adapt_array)
# Converts TEXT to np.array when selecting
sqlite3.register_converter("array", convert_array)
x = np.arange(12).reshape(2,6)
con = sqlite3.connect(":memory:", detect_types=sqlite3.PARSE_DECLTYPES)
cur = con.cursor()
cur.execute("create table test (arr array)")
With this setup, you can simply insert the NumPy array with no change in syntax:
cur.execute("insert into test (arr) values (?)", (x, ))
And retrieve the array directly from sqlite as a NumPy array:
cur.execute("select arr from test")
data = cur.fetchone()[0]
print(data)
# [[ 0 1 2 3 4 5]
# [ 6 7 8 9 10 11]]
print(type(data))
# <type 'numpy.ndarray'>
Happy Leap Second has it close but I kept getting an automatic casting to string.
Also if you check out this other post: a fun debate on using buffer or Binary to push non text data into sqlite you see that the documented approach is to avoid the buffer all together and use this chunk of code.
def adapt_array(arr):
out = io.BytesIO()
np.save(out, arr)
out.seek(0)
return sqlite3.Binary(out.read())
I haven’t heavily tested this in python 3, but it seems to work in python 2.7
I think that matlab format is a really convenient way to store and retrieve numpy arrays. Is really fast and the disk and memory footprint is quite the same.
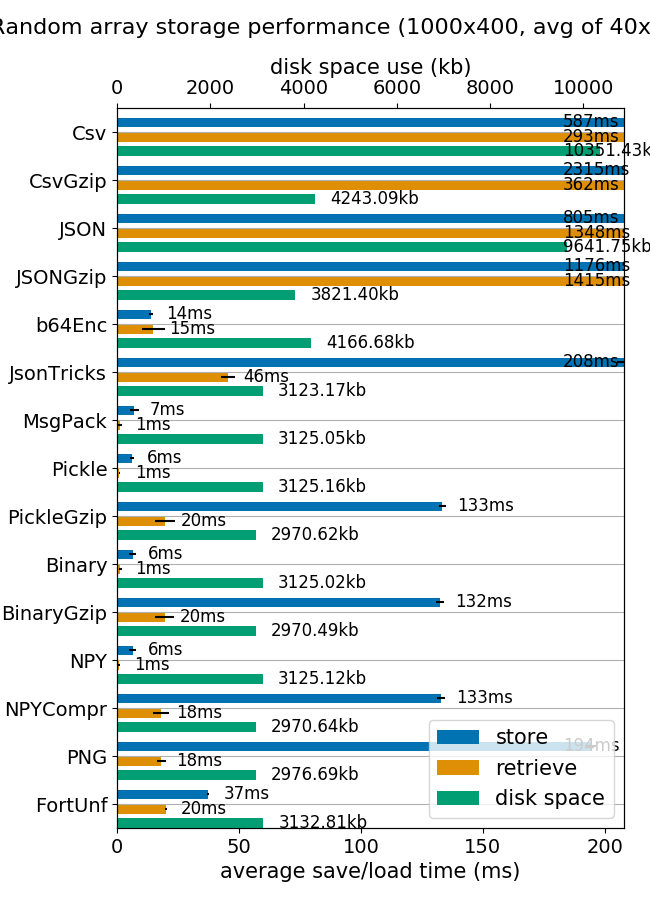
(image from mverleg benchmarks)
But if for any reason you need to store the numpy arrays into SQLite I suggest to add some compression capabilities.
The extra lines from unutbu code is pretty simple
compressor = 'zlib' # zlib, bz2
def adapt_array(arr):
"""
http://stackoverflow.com/a/31312102/190597 (SoulNibbler)
"""
# zlib uses similar disk size that Matlab v5 .mat files
# bz2 compress 4 times zlib, but storing process is 20 times slower.
out = io.BytesIO()
np.save(out, arr)
out.seek(0)
return sqlite3.Binary(out.read().encode(compressor)) # zlib, bz2
def convert_array(text):
out = io.BytesIO(text)
out.seek(0)
out = io.BytesIO(out.read().decode(compressor))
return np.load(out)
The results testing with MNIST database gives were:
$ ./test_MNIST.py
[69900]: 99% remain: 0 secs
Storing 70000 images in 379.9 secs
Retrieve 6990 images in 9.5 secs
$ ls -lh example.db
-rw-r--r-- 1 agp agp 69M sep 22 07:27 example.db
$ ls -lh mnist-original.mat
-rw-r--r-- 1 agp agp 53M sep 20 17:59 mnist-original.mat
```
using zlib, and
$ ./test_MNIST.py
[69900]: 99% remain: 12 secs
Storing 70000 images in 8536.2 secs
Retrieve 6990 images in 37.4 secs
$ ls -lh example.db
-rw-r--r-- 1 agp agp 19M sep 22 03:33 example.db
$ ls -lh mnist-original.mat
-rw-r--r-- 1 agp agp 53M sep 20 17:59 mnist-original.mat
using bz2
Comparing Matlab V5 format with bz2 on SQLite, the bz2 compression is around 2.8, but the access time is quite long comparing to Matlab format (almost instantaneously vs more than 30 secs). Maybe is worthy only for really huge databases where the learning process is much time consuming than access time or where the database footprint is needed to be as small as possible.
Finally note that bipz/zlib ratio is around 3.7 and zlib/matlab requires 30% more space.
The full code if you want to play yourself is:
import sqlite3
import numpy as np
import io
compressor = 'zlib' # zlib, bz2
def adapt_array(arr):
"""
http://stackoverflow.com/a/31312102/190597 (SoulNibbler)
"""
# zlib uses similar disk size that Matlab v5 .mat files
# bz2 compress 4 times zlib, but storing process is 20 times slower.
out = io.BytesIO()
np.save(out, arr)
out.seek(0)
return sqlite3.Binary(out.read().encode(compressor)) # zlib, bz2
def convert_array(text):
out = io.BytesIO(text)
out.seek(0)
out = io.BytesIO(out.read().decode(compressor))
return np.load(out)
sqlite3.register_adapter(np.ndarray, adapt_array)
sqlite3.register_converter("array", convert_array)
dbname = 'example.db'
def test_save_sqlite_arrays():
"Load MNIST database (70000 samples) and store in a compressed SQLite db"
os.path.exists(dbname) and os.unlink(dbname)
con = sqlite3.connect(dbname, detect_types=sqlite3.PARSE_DECLTYPES)
cur = con.cursor()
cur.execute("create table test (idx integer primary key, X array, y integer );")
mnist = fetch_mldata('MNIST original')
X, y = mnist.data, mnist.target
m = X.shape[0]
t0 = time.time()
for i, x in enumerate(X):
cur.execute("insert into test (idx, X, y) values (?,?,?)",
(i, y, int(y[i])))
if not i % 100 and i > 0:
elapsed = time.time() - t0
remain = float(m - i) / i * elapsed
print "r[%5d]: %3d%% remain: %d secs" % (i, 100 * i / m, remain),
sys.stdout.flush()
con.commit()
con.close()
elapsed = time.time() - t0
print
print "Storing %d images in %0.1f secs" % (m, elapsed)
def test_load_sqlite_arrays():
"Query MNIST SQLite database and load some samples"
con = sqlite3.connect(dbname, detect_types=sqlite3.PARSE_DECLTYPES)
cur = con.cursor()
# select all images labeled as '2'
t0 = time.time()
cur.execute('select idx, X, y from test where y = 2')
data = cur.fetchall()
elapsed = time.time() - t0
print "Retrieve %d images in %0.1f secs" % (len(data), elapsed)
if __name__ == '__main__':
test_save_sqlite_arrays()
test_load_sqlite_arrays()
The other methods specified didn’t work for me. And well there seems to be a numpy.tobytes method now and a numpy.fromstring (which works on byte strings) but is deprecated and the recommended method is numpy.frombuffer.
import sqlite3
import numpy as np
sqlite3.register_adapter(np.array, lambda arr: arr.tobytes())
sqlite3.register_converter("array", np.frombuffer)
I’ve tested it in my application and it works well for me on Python 3.7.3 and numpy 1.16.2
numpy.fromstring gives the same outputs along with DeprecationWarning: The binary mode of fromstring is deprecated, as it behaves surprisingly on unicode inputs. Use frombuffer instead
Ready to use code based on @unutbu’s answer (cleaned a bit, no need to seek, etc.), and test with a 2D ndarray:
import sqlite3, numpy as np, io
def adapt_array(arr):
out = io.BytesIO()
np.save(out, arr)
return sqlite3.Binary(out.getvalue())
sqlite3.register_adapter(np.ndarray, adapt_array)
sqlite3.register_converter("array", lambda x: np.load(io.BytesIO(x)))
x = np.random.rand(100, 100)
con = sqlite3.connect(":memory:", detect_types=sqlite3.PARSE_DECLTYPES)
con.execute("create table test (arr array)")
con.execute("insert into test (arr) values (?)", (x, ))
for r in con.execute("select arr from test"):
print(r[0])
You can use this (see @gavin’s answer) instead if and only if you only work with 1D arrays:
sqlite3.register_adapter(np.ndarray, lambda arr: arr.tobytes())
sqlite3.register_converter("array", np.frombuffer)
I’m trying to store a numpy array of about 1000 floats in a sqlite3 database but I keep getting the error “InterfaceError: Error binding parameter 1 – probably unsupported type”.
I was under the impression a BLOB data type could be anything but it definitely doesn’t work with a numpy array. Here’s what I tried:
import sqlite3 as sql
import numpy as np
con = sql.connect('test.bd',isolation_level=None)
cur = con.cursor()
cur.execute("CREATE TABLE foobar (id INTEGER PRIMARY KEY, array BLOB)")
cur.execute("INSERT INTO foobar VALUES (?,?)", (None,np.arange(0,500,0.5)))
con.commit()
Is there another module I can use to get the numpy array into the table? Or can I convert the numpy array into another form in Python (like a list or string I can split) that sqlite will accept? Performance isn’t a priority. I just want it to work!
Thanks!
This works for me:
import sqlite3 as sql
import numpy as np
import json
con = sql.connect('test.db',isolation_level=None)
cur = con.cursor()
cur.execute("DROP TABLE FOOBAR")
cur.execute("CREATE TABLE foobar (id INTEGER PRIMARY KEY, array BLOB)")
cur.execute("INSERT INTO foobar VALUES (?,?)", (None, json.dumps(np.arange(0,500,0.5).tolist())))
con.commit()
cur.execute("SELECT * FROM FOOBAR")
data = cur.fetchall()
print data
data = cur.fetchall()
my_list = json.loads(data[0][1])
You could register a new array data type with sqlite3:
import sqlite3
import numpy as np
import io
def adapt_array(arr):
"""
http://stackoverflow.com/a/31312102/190597 (SoulNibbler)
"""
out = io.BytesIO()
np.save(out, arr)
out.seek(0)
return sqlite3.Binary(out.read())
def convert_array(text):
out = io.BytesIO(text)
out.seek(0)
return np.load(out)
# Converts np.array to TEXT when inserting
sqlite3.register_adapter(np.ndarray, adapt_array)
# Converts TEXT to np.array when selecting
sqlite3.register_converter("array", convert_array)
x = np.arange(12).reshape(2,6)
con = sqlite3.connect(":memory:", detect_types=sqlite3.PARSE_DECLTYPES)
cur = con.cursor()
cur.execute("create table test (arr array)")
With this setup, you can simply insert the NumPy array with no change in syntax:
cur.execute("insert into test (arr) values (?)", (x, ))
And retrieve the array directly from sqlite as a NumPy array:
cur.execute("select arr from test")
data = cur.fetchone()[0]
print(data)
# [[ 0 1 2 3 4 5]
# [ 6 7 8 9 10 11]]
print(type(data))
# <type 'numpy.ndarray'>
Happy Leap Second has it close but I kept getting an automatic casting to string.
Also if you check out this other post: a fun debate on using buffer or Binary to push non text data into sqlite you see that the documented approach is to avoid the buffer all together and use this chunk of code.
def adapt_array(arr):
out = io.BytesIO()
np.save(out, arr)
out.seek(0)
return sqlite3.Binary(out.read())
I haven’t heavily tested this in python 3, but it seems to work in python 2.7
I think that matlab format is a really convenient way to store and retrieve numpy arrays. Is really fast and the disk and memory footprint is quite the same.

(image from mverleg benchmarks)
But if for any reason you need to store the numpy arrays into SQLite I suggest to add some compression capabilities.
The extra lines from unutbu code is pretty simple
compressor = 'zlib' # zlib, bz2
def adapt_array(arr):
"""
http://stackoverflow.com/a/31312102/190597 (SoulNibbler)
"""
# zlib uses similar disk size that Matlab v5 .mat files
# bz2 compress 4 times zlib, but storing process is 20 times slower.
out = io.BytesIO()
np.save(out, arr)
out.seek(0)
return sqlite3.Binary(out.read().encode(compressor)) # zlib, bz2
def convert_array(text):
out = io.BytesIO(text)
out.seek(0)
out = io.BytesIO(out.read().decode(compressor))
return np.load(out)
The results testing with MNIST database gives were:
$ ./test_MNIST.py
[69900]: 99% remain: 0 secs
Storing 70000 images in 379.9 secs
Retrieve 6990 images in 9.5 secs
$ ls -lh example.db
-rw-r--r-- 1 agp agp 69M sep 22 07:27 example.db
$ ls -lh mnist-original.mat
-rw-r--r-- 1 agp agp 53M sep 20 17:59 mnist-original.mat
```
using zlib, and
$ ./test_MNIST.py
[69900]: 99% remain: 12 secs
Storing 70000 images in 8536.2 secs
Retrieve 6990 images in 37.4 secs
$ ls -lh example.db
-rw-r--r-- 1 agp agp 19M sep 22 03:33 example.db
$ ls -lh mnist-original.mat
-rw-r--r-- 1 agp agp 53M sep 20 17:59 mnist-original.mat
using bz2
Comparing Matlab V5 format with bz2 on SQLite, the bz2 compression is around 2.8, but the access time is quite long comparing to Matlab format (almost instantaneously vs more than 30 secs). Maybe is worthy only for really huge databases where the learning process is much time consuming than access time or where the database footprint is needed to be as small as possible.
Finally note that bipz/zlib ratio is around 3.7 and zlib/matlab requires 30% more space.
The full code if you want to play yourself is:
import sqlite3
import numpy as np
import io
compressor = 'zlib' # zlib, bz2
def adapt_array(arr):
"""
http://stackoverflow.com/a/31312102/190597 (SoulNibbler)
"""
# zlib uses similar disk size that Matlab v5 .mat files
# bz2 compress 4 times zlib, but storing process is 20 times slower.
out = io.BytesIO()
np.save(out, arr)
out.seek(0)
return sqlite3.Binary(out.read().encode(compressor)) # zlib, bz2
def convert_array(text):
out = io.BytesIO(text)
out.seek(0)
out = io.BytesIO(out.read().decode(compressor))
return np.load(out)
sqlite3.register_adapter(np.ndarray, adapt_array)
sqlite3.register_converter("array", convert_array)
dbname = 'example.db'
def test_save_sqlite_arrays():
"Load MNIST database (70000 samples) and store in a compressed SQLite db"
os.path.exists(dbname) and os.unlink(dbname)
con = sqlite3.connect(dbname, detect_types=sqlite3.PARSE_DECLTYPES)
cur = con.cursor()
cur.execute("create table test (idx integer primary key, X array, y integer );")
mnist = fetch_mldata('MNIST original')
X, y = mnist.data, mnist.target
m = X.shape[0]
t0 = time.time()
for i, x in enumerate(X):
cur.execute("insert into test (idx, X, y) values (?,?,?)",
(i, y, int(y[i])))
if not i % 100 and i > 0:
elapsed = time.time() - t0
remain = float(m - i) / i * elapsed
print "r[%5d]: %3d%% remain: %d secs" % (i, 100 * i / m, remain),
sys.stdout.flush()
con.commit()
con.close()
elapsed = time.time() - t0
print
print "Storing %d images in %0.1f secs" % (m, elapsed)
def test_load_sqlite_arrays():
"Query MNIST SQLite database and load some samples"
con = sqlite3.connect(dbname, detect_types=sqlite3.PARSE_DECLTYPES)
cur = con.cursor()
# select all images labeled as '2'
t0 = time.time()
cur.execute('select idx, X, y from test where y = 2')
data = cur.fetchall()
elapsed = time.time() - t0
print "Retrieve %d images in %0.1f secs" % (len(data), elapsed)
if __name__ == '__main__':
test_save_sqlite_arrays()
test_load_sqlite_arrays()
The other methods specified didn’t work for me. And well there seems to be a numpy.tobytes method now and a numpy.fromstring (which works on byte strings) but is deprecated and the recommended method is numpy.frombuffer.
import sqlite3
import numpy as np
sqlite3.register_adapter(np.array, lambda arr: arr.tobytes())
sqlite3.register_converter("array", np.frombuffer)
I’ve tested it in my application and it works well for me on Python 3.7.3 and numpy 1.16.2
numpy.fromstring gives the same outputs along with DeprecationWarning: The binary mode of fromstring is deprecated, as it behaves surprisingly on unicode inputs. Use frombuffer instead
Ready to use code based on @unutbu’s answer (cleaned a bit, no need to seek, etc.), and test with a 2D ndarray:
import sqlite3, numpy as np, io
def adapt_array(arr):
out = io.BytesIO()
np.save(out, arr)
return sqlite3.Binary(out.getvalue())
sqlite3.register_adapter(np.ndarray, adapt_array)
sqlite3.register_converter("array", lambda x: np.load(io.BytesIO(x)))
x = np.random.rand(100, 100)
con = sqlite3.connect(":memory:", detect_types=sqlite3.PARSE_DECLTYPES)
con.execute("create table test (arr array)")
con.execute("insert into test (arr) values (?)", (x, ))
for r in con.execute("select arr from test"):
print(r[0])
You can use this (see @gavin’s answer) instead if and only if you only work with 1D arrays:
sqlite3.register_adapter(np.ndarray, lambda arr: arr.tobytes())
sqlite3.register_converter("array", np.frombuffer)